Machine Learning
Machine learning and deep learning have revolutionized the way we approach scientific research and problem-solving. At MTEC, we use this powerful technology to address some of the toughest medical and clinical challenges facing society today. With the help of advanced GPUs and specialized software frameworks, we are able to conduct cutting-edge research in a variety of medical domains, including brain anomaly detection, paranasal anomaly detection, polyp segmentation from colonoscopy videos, skin lesion classification, and the analysis of multi-dimensional medical data over time. Join us at MTEC and become a part of this exciting field where we continue to push the boundaries of what is possible with Deep Learning.
Selected Publications
- F. Behrendt, M. Bengs, D. Bhattacharya, J. Krüger, R. Opfer, A. Schlaefer (2023). A systematic approach to deep learning-based nodule detection in chest radiographs. Scientific Reports. 13. (1), 10120 [Abstract]
[doi][www][BibTex]
- D. Bhattacharya, B. T. Becker, F. Behrendt, M. Bengs, D. Beyersdorff, D. Eggert, E. Petersen, F. Jansen, M. Petersen, B. Cheng, C. Betz, A. Schlaefer, A. S. Hoffmann (2022). Supervised Contrastive Learning to Classify Paranasal Anomalies in the Maxillary Sinus. In Wang, Linwei, Dou, Qi, Fletcher, P. Thomas, Speidel, Stefanie, Li, Shuo (Eds.) Medical Image Computing, Computer Assisted Intervention -- MICCAI 2022 Springer Nature Switzerland: Cham 429-438 [Abstract]
[doi][www][BibTex]
- F. Behrendt, M. Bengs, F. Rogge, J. Krüger, R. Opfer, A. Schlaefer (2022). Unsupervised Anomaly Detection in 3D Brain MRI Using Deep Learning with Impured Training Data. 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI) 1-4 [Abstract]
[doi][BibTex]
- M. Bengs, F. Behrendt, J. Krüger, R. Opfer, A. Schlaefer (2021). Three-dimensional deep learning with spatial erasing for unsupervised anomaly segmentation in brain MRI. International Journal of Computer Assisted Radiology and Surgery. 16. (9), 1413-1423 [Abstract]
[doi][www][BibTex]
-
N. Gessert, M. Bengs, M. Schlüter, A. Schlaefer
(2020).
Deep learning with 4D spatio-temporal data representations for OCT-based force estimation.
Medical Image Analysis.
64
(101730),
[Abstract][doi][www][BibTex]
-
N. Gessert, M. Nielsen, M. Shaikh, R. Werner, A. Schlaefer
(2020).
Skin lesion classification using ensembles of multi-resolution EfficientNets with meta data.
MethodsX.
7
100864.
[Abstract][doi][www][BibTex]
-
N. Gessert, M. Bengs, L. Wittig, D. Drömann, T. Keck, A. Schlaefer, D. B. Ellebrecht
(2019).
Deep transfer learning methods for colon cancer classification in confocal laser microscopy images.
International Journal of Computer Assisted Radiology and Surgery.14(11), 1837–1845.
[Abstract][doi][www][BibTex]
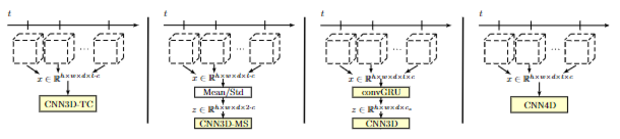
Spatio-Temporal Deep Learning Methods
Sequences of medical image data are at the heart of various clinical applications such as shear wave elastography, motion analysis, diseases progression or functional analysis, i.e. behavior or activity of organs over time. Within the field of medical imaging, there is a plethora of medical imaging modalities that can be used to this end. These modalities come with different temporal and spatial resolution and can range from one dimensional imaging over time up to three dimensional imaging over time.
In this context, we work on deep learning methods for spatio-temporal data analysis and use concepts such as spatio-temporal convolutional neural networks, recurrent neural networks and combined approaches. A major difference compared to conventional image processing methods is that our deep learning approaches process data in an end-to-end fashion without the required of pre-defined feature selection and extraction. While we address a wide range of applications and imaging modalities, we are particularly interested in data processing for real-time applications.
In previous works, we have addressed tasks such as autism spectrum disorder classification from functional magnetic resonance imaging, disease progression analysis from magnetic resonance imaging, motion analysis and force estimation from optical coherence tomopgrahy image volumes, and shear wave elastography from optical coherence tomopgrahy image volumes and ultrasound images. We also adapted our approaches to other imaging modalities such as hyperspectral imaging, where an image sequence at serval spectral bands is acquired. Here, we have demonstrated in-vivo cancer detection and classification adopting our deep learning concepts for spatio-temporal data processing.
Contrastive Learning and its use in medical imaging
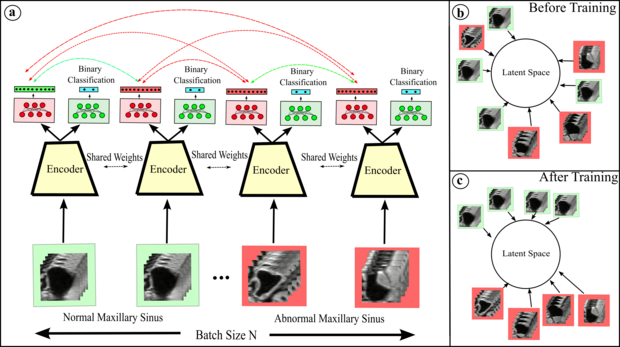
Contrastive learning is a powerful technique that allows an encoder to learn meaningful representations of data by mapping positive pairs close together in the embedding space and pushing negative pairs away. One popular method for implementing contrastive learning is SimCLR, in which an image is transformed through two different random transformations. These transformed "views" of the image are considered a positive pair, and every other image in the mini-batch is used as a negative pair for comparison. While augmentations are often used to improve the effectiveness of contrastive learning, it is important to be mindful of the potential for these transformations to negatively impact the quality of the learned representation. That's why at our institute, we are dedicated to researching novel positive and negative pair strategies that are tailored to the specific downstream task at hand, as well as developing ways to quantitatively analyze the representations learned by the network. As an example of our work, we have used contrastive learning to classify paranasal anomalies in the maxillary sinus and found that our method was highly data efficient and outperformed regular cross entropy trained models.
Self-Supervision in Medical Image Analysis
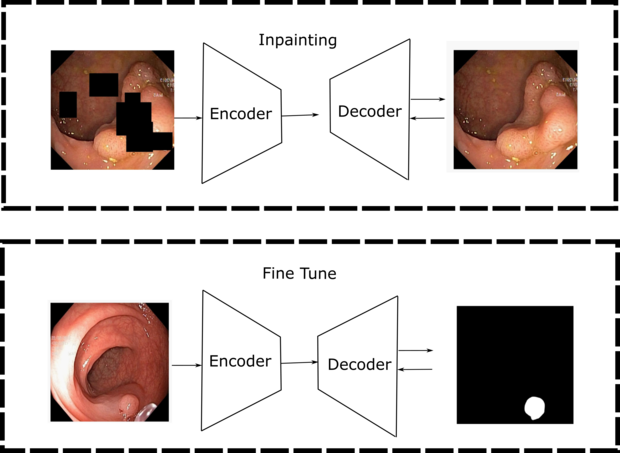
Obtaining labeled medical imaging data can be a challenging and time-consuming process, as it requires the expertise of trained clinicians to label individual samples in the dataset. This often leads to a situation where we have access to a small, labeled dataset and a larger, unlabeled dataset. However, self-supervised learning techniques offer a solution to this problem by allowing us to learn visual features from the unlabeled data. By generating supervisory signals through pretext tasks like colorization, jigsaw puzzles, inpainting, and contrastive learning, we can train neural networks without the need for extensive manual labeling. At our institute, we are dedicated to finding novel pretraining strategies that make use of unlabeled datasets to learn meaningful representations that can improve the performance of downstream tasks.
Unsupervised anomaly detection in Brain MRI scans
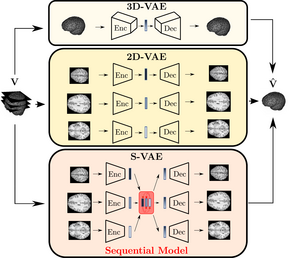
We work on deep learning-based unsupervised anomaly detection (UAD) methods to identify abnormalities in human brains without requiring an extensive amount of labeled training data. Thereby, we encode the appearance of healthy brains and detect anomalies as outliers from the learned latent representation. So far, this task has often been done in a slice-by-slice fashion, ignoring the 3D structure of the brain. Our work focuses on leveraging the meaningful 3D information, either end-to-end 3D processing or through efficient alternatives such as modeling inter-slice dependencies.
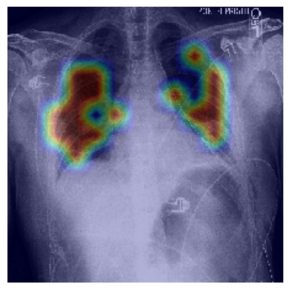
Automatic interpretation of chest x-rays
To develop a support tool for chest x-ray interpretation, we study deep learning-based classification and object detection algorithms. We investigate the application of very recent attention-based architectures such as vision transformers. To reduce the need for an excessive amount of labelled training data, our studies include efficient training strategies that utilize self-supervision, knowledge distillation or generated training data.
Contact
- Alexander Schlaefer (schlaefer(at)tuhh.de)
- Lennart Maack (lennart.maack(at)tuhh.de)
- Finn Behrendt (finn.behrendt(at)tuhh.de)
- Adrian Rudloff (adrian.rudloff(at)tuhh.de)